

11/25/2013
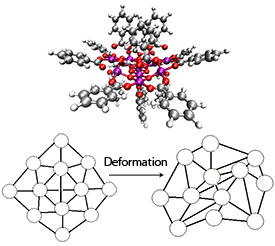
Reproduced from Ref. 1 © 2013 The Royal Society
Molecular magnets can be handled like classical particles owing to their relatively large size, but they also exhibit quantum magnetic properties thanks to their unpaired electron spins. This unique behavior makes them attractive as high-density information storage materials and for spintronic-based computing. Currently, chemists struggle to attach molecular magnets to device surfaces, however, because these particles have a tendency to warp and lose their magnetic properties on adsorption.
To overcome this difficulty, an international team led by Daniel Packwood from the AIMR at Tohoku University has now developed innovative ‘mathematical chemistry’ techniques that can predict novel molecular magnets with deformation-resistant magnetic moments1.
Most molecular magnets are made by linking transition metals, such as manganese and iron, into complex clusters through hydrocarbon or oxygen ‘bridges’ (see image). When the coupling interactions between the unpaired metal ion electrons within these metal–organic complexes are strong enough, the molecules acquire an appreciable magnetic moment. Packwood and colleagues hypothesized that the intimate relationship between structure and magnetic behavior in these systems could mean that certain geometric networks, currently unknown to chemists, exist with intrinsically stable spin properties.
To find these structures, the researchers devised an original model that examines how one physical property — the energy of the molecule’s ground spin state — changes with small, random shape deformations of a molecular magnet. “Creating a realistic model of a molecule is not so difficult,” says Packwood. “However, the key challenge is striking a good balance between physical realism and sufficient simplicity for mathematical treatment.”
The team’s analysis showed that two-dimensional molecules containing a sufficient number of spin centers can possess extraordinarily stable ‘weak topological invariant’ magnetic moments. “When mathematicians talk about topological invariants, they usually mean properties that don’t change under arbitrary stretching or squeezing deformations,” explains Packwood. “A weak topological invariant is similar; however, it can change under deformations that have a very low probability of occurring. This difference significantly broadens the applicability of the topological invariance concept to studying real systems.”
Surprisingly, the researchers’ calculations showed that only between 20 and 50 spin centers were needed for a two-dimensional molecule to achieve robust and stable magnetism. They also found that resistance to shape deformations improved when the number of ring-shaped networks in the model was kept to a minimum. As the structures predicted by this joint mathematics–materials science study are well within the size range of existing molecular magnets, Packwood and colleagues are confident that they represent practical targets for future chemical synthetic efforts.
Packwood, D. M., Reaves, K. T., Federici, F. L., Katzgraber, H. G. & Teizer, W. Two-dimensional molecular magnets with weak topological invariant magnetic moments: Mathematical prediction of targets for chemical synthesis. Proceedings of the Royal Society A 469, 20130373 (2013). | article
This research highlight has been approved by the authors of the original article and all information and data contained within has been provided by said authors.